Another collections of sequences with bioinformatics tools can be found at http://www.flugenome.org, but this resource is not updated with the current data and can’t be used for genotyping of the latest sequences. Check also GISAID Platform at: http://platform.gisaid.org/
Bioinformatics of Swine Flu
April 28, 2009 at 11:07 pm 4 comments
The swine influenza A (H1N1) virus is an RNA virus coding for 8 genes. The sequences of these genes from strains isolated in Texas, California, New York and Kansas can be downloaded from GenBank:
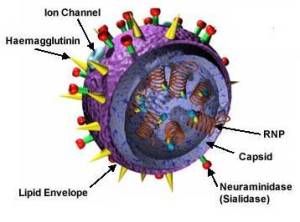
- HA encodes hemagglutinin (about 500 molecules of hemagglutinin are needed to make one virion). This protein determines the extent of infection into host organism, bronchial epithelial cells, lungs and other organs.
- NA encodes neuraminidase (about 100 molecules of neuraminidase are needed to make one virus particle).
- NP encodes nucleoprotein.
- M encodes two matrix proteins (the M1 and the M2, which is an ion channel) by using different reading frames from the same RNA segment (about 3000 matrix protein molecules are needed to make one virion).
- NS encodes two distinct non-structural proteins (NS1 and NEP) also by using different reading frames from the same RNA segment.
- PA encodes an RNA polymerase.
- PB1 encodes an RNA polymerase and PB1-F2 protein (induces apoptosis) by using different reading frames from the same RNA segment.
- PB2 encodes an RNA polymerase.
The genome is a composite of avian flu, human flu Type A, human flu Type B, Asian swine flu, and European swine flu. A strange combination having less than 0.1% chance of being a natural event. Most of the genes, including the hemagglutinin (HA) gene, are of the same family as seen in US pigs in the last decade, but the neuraminidase (NA) and matrix (M) protein genes are more like swine flu viruses seen in Eurasia. Other chunks of the genome came from influenza viruses carried by birds and humans. Novel swine flu isolates are resistant to the older antiviral adamantane class of drugs (M2 inhibitors), but sensitive to the adamantanes. Two anti-viral drugs on the market – Tamiflu and Relenza –can lessen the symptoms of swine flu.
Take a look at the BLAST results of 8 NA sequences coding for the protein targeted by drugs oseltamivir (Tamiflu) and zanamivir (Relenza). Perfect alignment to european swine flu sequences, minimal variations. The bird viruses come second best. Taking into account that bird flu easily developed resistance to the above mentioned drugs, and that vaccine development will be a challenge, this does not sound too good.
 Flu viruses are growing resistant to key weapon Tamiflu This year, Tamiflu resistance in that class of viruses has reached almost 100%, turning the tables on a drug designed to defeat resistance. A CDC team tested 1,155 A/H1N1 viruses from 45 states. They found that 142 viruses from 24 states were resistant to Tamiflu, or 12.3%. Earlier this year, 264 of 268 viruses tested were Tamiflu-resistant, or 98.5%, reported in The Journal of the American Medical Association.
Flu viruses are growing resistant to key weapon Tamiflu This year, Tamiflu resistance in that class of viruses has reached almost 100%, turning the tables on a drug designed to defeat resistance. A CDC team tested 1,155 A/H1N1 viruses from 45 states. They found that 142 viruses from 24 states were resistant to Tamiflu, or 12.3%. Earlier this year, 264 of 268 viruses tested were Tamiflu-resistant, or 98.5%, reported in The Journal of the American Medical Association.
CDC expects we will see more deaths. From a historical perspective, this is, indeed, scary: Asian Flu pandemic of 1957 and the Hong Kong Flu pandemic of 1968 occurred as a result of influenza mutated by antigenic shift, recombination similar to the one observed for current swine flu outbreak. Missense mutations account for other milder epidemics (1962, 1964, etc). The human swine flu outbreak continues to grow in the United States and internationally. So is it going to be a Worldwide Pandemic or a Case of the Sniffles? Let’s hope for the best, but prepare for the worst.
Related articles by Zemanta
- Swine Flu Unlikely to Affect the Economy (time.com)
- Swine Flu: Facts, Myths and Whether to Worry (abcnews.go.com)
- Swine Flu: What You Need to Know (abcnews.go.com)
- Resistance to Tamiflu Growing (nlm.nih.gov)
- Swine Flu: Is this the Big One? (grantlawrence.blogspot.com)
- Questions, answers about swine flu (seattletimes.nwsource.com)
- Why the hullabaloo about Swine Flu? (thegenesherpa.blogspot.com)
- CDC Readies Swine Flu Vaccine (time.com)
Entry filed under: Microworld. Tags: alignment, Avian influenza, bioinformatics, California, drug resistance, GenBank, Health, Infectious disease, Influenza, Oseltamivir, sequence, Swine influenza, US, Virus, Zanamivir.
4 Comments Add your own
Leave a reply to Bioinformatics of Swine Flu Cancel reply
Trackback this post | Subscribe to the comments via RSS Feed


1. Bioinformatics of Swine Flu | Health @ U Want 2 Know .Info | April 28, 2009 at 11:25 pm
[…] Original post by aurametrix […]
2. Bioinformatics of Swine Flu | April 29, 2009 at 12:29 am
[…] Original post by aurametrix […]
3. orespiple | May 7, 2009 at 2:43 pm
orespiple | May 7, 2009 at 2:43 pm
Hello
Tamiflu (oseltamivir) is successful in helping treat the virus if taken within several days of having symptoms, according to the Centers for Disease Control and Prevention. The drugs can shorten the length of the disease and its severity
Where to find tamiflu?- https://twitter.com/Swine_Flu_Treat
4. black hattitude | October 21, 2009 at 7:19 pm
black hattitude | October 21, 2009 at 7:19 pm
Hi,
thanks for the great quality of your blog, each time i come here, i’m amazed.
black hattitude.